
Next: Continuous action spaces Up: 8.5.2 Dynamic Programming with Previous: 8.5.2 Dynamic Programming with
Consider a problem formulation that is identical to Formulation
8.1 except that ![]() is allowed to be continuous. Assume
that
is allowed to be continuous. Assume
that ![]() is bounded, and assume for now that the action space,
is bounded, and assume for now that the action space, ![]() ,
it finite for all
,
it finite for all ![]() . Backward value iteration can be applied.
The dynamic programming arguments and derivation are identical to
those in Section 2.3. The resulting recurrence is
identical to (2.11) and is repeated here for convenience:
. Backward value iteration can be applied.
The dynamic programming arguments and derivation are identical to
those in Section 2.3. The resulting recurrence is
identical to (2.11) and is repeated here for convenience:
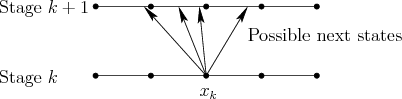 |
Suppose that a finite set
![]() of samples is used to
represent cost-to-go functions over
of samples is used to
represent cost-to-go functions over ![]() . The evaluation of
(8.56) using interpolation is depicted in Figure
8.19. In general, the samples should be chosen to reduce
the dispersion (defined in Section
5.2.3) as much as possible. This prevents attempts to
approximate the cost-to-go function on large areas that contain no
sample points. The rate of convergence ultimately depends on the
dispersion [92] (in combination with Lipschitz conditions on
the state transition equation and the cost functional). To simplify
notation and some other issues, assume that
. The evaluation of
(8.56) using interpolation is depicted in Figure
8.19. In general, the samples should be chosen to reduce
the dispersion (defined in Section
5.2.3) as much as possible. This prevents attempts to
approximate the cost-to-go function on large areas that contain no
sample points. The rate of convergence ultimately depends on the
dispersion [92] (in combination with Lipschitz conditions on
the state transition equation and the cost functional). To simplify
notation and some other issues, assume that ![]() is a grid of regularly
spaced points in
is a grid of regularly
spaced points in
![]() .
.
First, consider the case in which
![]() . Let
. Let
![]() , in which
, in which ![]() . For example, if
. For example, if ![]() , then
, then
![]() . Note that this always yields points
on the boundary of
. Note that this always yields points
on the boundary of ![]() , which ensures that for any point in
, which ensures that for any point in ![]() there are samples both above and below it. Let
there are samples both above and below it. Let ![]() be the largest
integer such that
be the largest
integer such that ![]() . This implies that
. This implies that
![]() . The
samples
. The
samples ![]() and
and ![]() are called interpolation neighbors
of
are called interpolation neighbors
of ![]() .
.
The value of ![]() in (8.56) at any
in (8.56) at any
![]() can be obtained via linear interpolation as
can be obtained via linear interpolation as
The interpolation idea can be naturally extended to multiple
dimensions. Let ![]() be a bounded subset of
be a bounded subset of
![]() . Let
. Let ![]() represent an
represent an ![]() -dimensional grid of points in
-dimensional grid of points in
![]() . Each sample
in
. Each sample
in ![]() is denoted by
is denoted by
![]() . For some
. For some ![]() ,
there are
,
there are ![]() interpolation neighbors that ``surround'' it. These
are the corners of an
interpolation neighbors that ``surround'' it. These
are the corners of an ![]() -dimensional cube that contains
-dimensional cube that contains ![]() . Let
. Let
![]() . Let
. Let ![]() denote the largest integer for which
the
denote the largest integer for which
the ![]() th coordinate of
th coordinate of
![]() is less than
is less than ![]() .
The
.
The ![]() samples are all those for which either
samples are all those for which either ![]() or
or ![]() appears in the expression
appears in the expression
![]() , for each
, for each
![]() . This requires that samples exist in
. This requires that samples exist in ![]() for all
of these cases. Note that
for all
of these cases. Note that ![]() may be a complicated subset of
may be a complicated subset of
![]() ,
provided that for any
,
provided that for any ![]() , all of the required
, all of the required ![]() interpolation neighbors are in
interpolation neighbors are in ![]() . Using the
. Using the ![]() interpolation
neighbors, the value of
interpolation
neighbors, the value of ![]() in (8.56) on any
in (8.56) on any ![]() can be obtained via multi-linear interpolation. In the
case of
can be obtained via multi-linear interpolation. In the
case of ![]() , this is expressed as
, this is expressed as
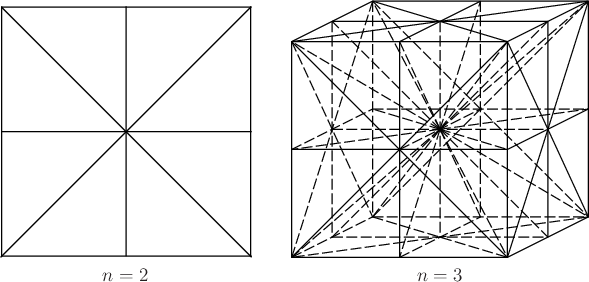 |
Unfortunately, the number of interpolation neighbors grows
exponentially with the dimension, ![]() . Instead of using all
. Instead of using all ![]() interpolation neighbors, one improvement is to decompose the cube
defined by the
interpolation neighbors, one improvement is to decompose the cube
defined by the ![]() samples into simplexes. Each simplex has only
samples into simplexes. Each simplex has only
![]() samples as its vertices. Only the vertices of the simplex that
contains
samples as its vertices. Only the vertices of the simplex that
contains ![]() are declared to be the interpolation neighbors of
are declared to be the interpolation neighbors of ![]() ;
this reduces the cost of evaluating
;
this reduces the cost of evaluating
![]() to
to ![]() time.
The problem, however, is that determining the simplex that contains
time.
The problem, however, is that determining the simplex that contains
![]() may be a challenging point-location problem (a common
problem in computational geometry [264]). If
barycentric subdivision is used to decompose the cube using the
midpoints of all faces, then the point-location problem can be solved
in
may be a challenging point-location problem (a common
problem in computational geometry [264]). If
barycentric subdivision is used to decompose the cube using the
midpoints of all faces, then the point-location problem can be solved
in
![]() time [263,607,721], which is an
improvement over the
time [263,607,721], which is an
improvement over the ![]() scheme described above. Examples of
this decomposition are shown for two and three dimensions in Figure
8.20. This is sometimes called the
Coxeter-Freudenthal-Kuhn triangulation. Even though
scheme described above. Examples of
this decomposition are shown for two and three dimensions in Figure
8.20. This is sometimes called the
Coxeter-Freudenthal-Kuhn triangulation. Even though ![]() is not
too large due to practical performance considerations (typically,
is not
too large due to practical performance considerations (typically, ![]() ), substantial savings occur in implementations, even for
), substantial savings occur in implementations, even for ![]() .
.
It will be convenient to refer directly to the set of all points in
![]() for which all required interpolation neighbors exist. For any
finite set
for which all required interpolation neighbors exist. For any
finite set
![]() of sample points, let the interpolation region
of sample points, let the interpolation region ![]() be the set of all
be the set of all
![]() for which
for which
![]() can be computed by interpolation. This means that
can be computed by interpolation. This means that
![]() if and only if all interpolation neighbors of
if and only if all interpolation neighbors of ![]() lie in
lie in ![]() .
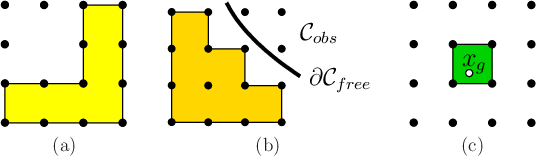
Figure 8.21a shows an example. Note that some sample
points may not contribute any points to
.
Figure 8.21a shows an example. Note that some sample
points may not contribute any points to ![]() . If a grid of samples is
used to approximate
. If a grid of samples is
used to approximate ![]() , then the volume of
, then the volume of
![]() approaches zero as the sampling resolution increases.
approaches zero as the sampling resolution increases.
 |