
Next: 7.6 Coverage Planning Up: 7.5 Folding Problems in Previous: Drug design
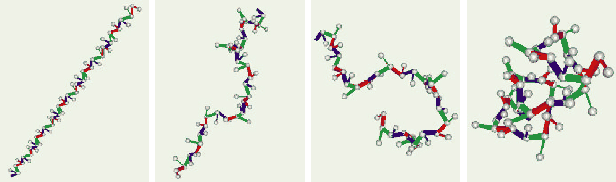
In computational biology, the problem of protein folding shares many similarities with drug design in that the molecules have rotatable bonds and energy functions are used to express good configurations. The problems are much more complicated, however, because the protein molecules are generally much larger than drug molecules. Instead of a dozen degrees of freedom, which is typical for a drug molecule, proteins have hundreds or thousands of degrees of freedom. When proteins appear in nature, they are usually in a folded, low-energy configuration. The structure problem involves determining precisely how the protein is folded so that its biological activity can be completely understood. In some studies, biologists are even interested in the pathway that a protein takes to arrive in its folded state [24,25]. This leads directly to an extension of motion planning that involves arriving at a goal state in which the molecule is folded. In [24,25], sampling-based planning algorithms were applied to compute folding pathways for proteins. The protein starts in an unfolded configuration and must arrive in a specified folded configuration without violating energy constraints along the way. Figure 7.34 shows an example from [25]. That work also draws interesting connections between protein folding and box folding, which was covered previously.
 |
Steven M LaValle 2020-08-14